 Ongoing projects: 1. I am using the expression of the Ret proto-oncogene in patches of cells in living developing tissue to recreate the early steps in cancer progression in order to find new ways by which cancer cells manage to survive, grow and escape from the site of origin. Steps leading up to metastasis. This is in the hope to discover new ways in which cancer progression can be blocked.
Ongoing projects: 1. I am using the expression of the Ret proto-oncogene in patches of cells in living developing tissue to recreate the early steps in cancer progression in order to find new ways by which cancer cells manage to survive, grow and escape from the site of origin. Steps leading up to metastasis. This is in the hope to discover new ways in which cancer progression can be blocked.
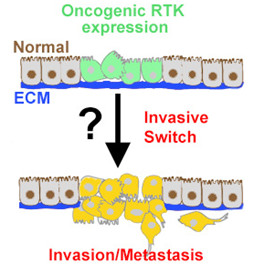
I am most interested in finding factors important for cancer spreading. What drives cancer cells out of their environment? Observations (ours, unpublished and others) suggest that the hostile nature of the microenvironment surrounding the early tumour cells, which can be nutrient and oxygen-poor can trigger changes that activate cell migration. One of the projects in the lab aims to investigate the effects of changes in nutrient driven signalling pathways to tumour cells tendency to migrate.
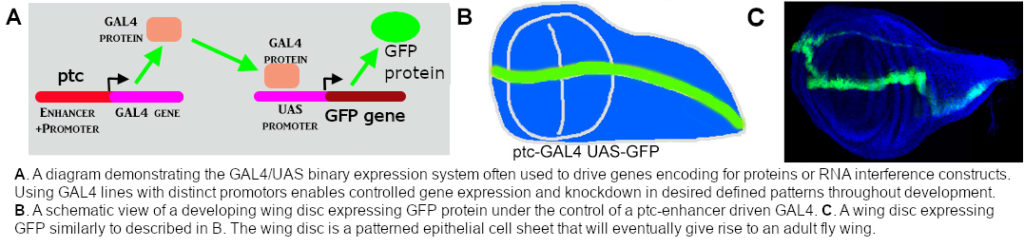
We use the GAL4/UAS binary expression system to simultaneously express oncogeneic Ret and RNAi interference constructs (knockdown of gene function) in a patch of cells of the epithelial sheet that makes the fly wing. Using this approach, or combining it with Pharmacological treatments, we can see which genes and processes are required for the spreading/migration of Ret oncogene expressing cells. An example of this is shown below. Genetic or pharmacological inhibition of Factor B leads to an increased spread of GFP-positive tumour cells. 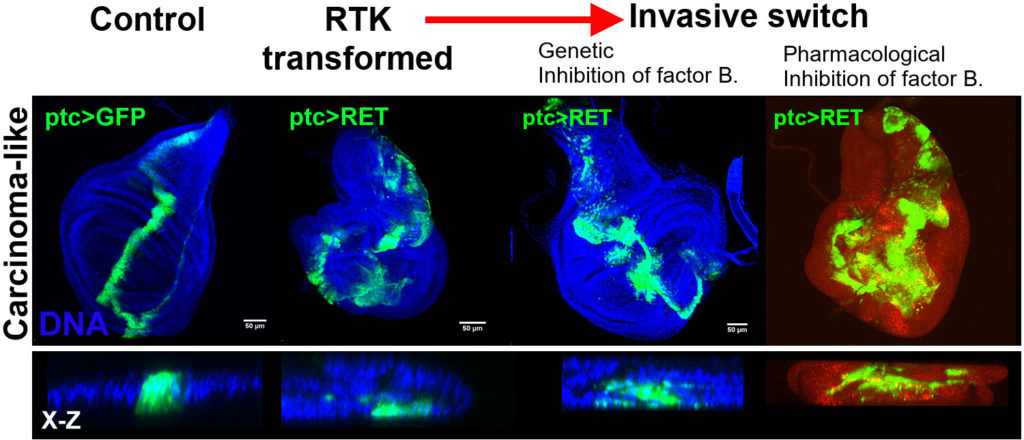
Project 2. Are signals from the neighbouring normal cells involved in the spreading of Ret positive cells? A non-autonomous (NA) effect. Can we stop this or make the elimination/stop the spread of cancer cells by normal cells more effective? To address this the fly model can serve us well. With the controlled expression of an oncogene in a patch of cells coupled to the ability to disrupt gene function in the surrounding cells, we can now ask which genes within the tumour contribute to its spread, and which genes in the surrounding tissue are needed to resist the spread or invasion by the tumour cells. 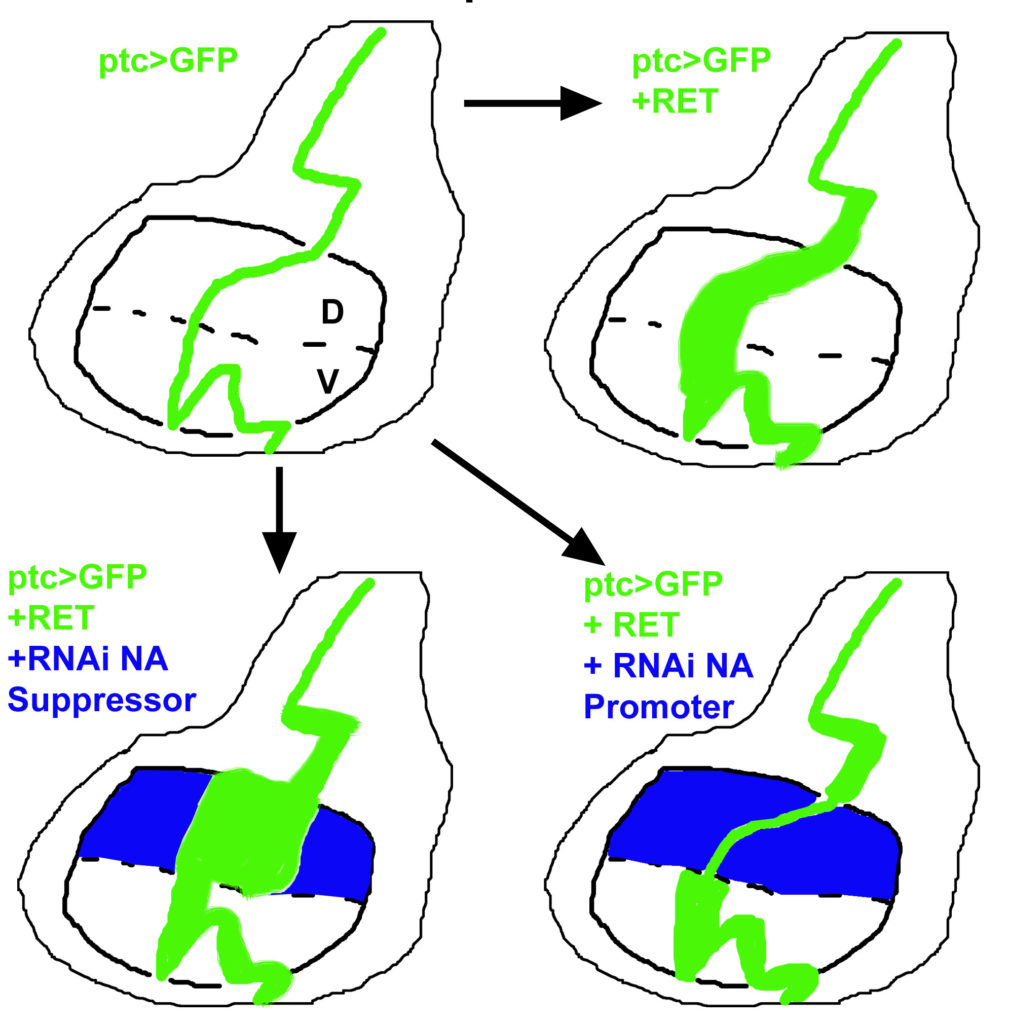
Identifying genes and the pathways in which they act in normal cells that surround a tumour that can help eliminate tumour spreading provide a new approach to cancer prevention. Potentially the activity of these pathways can be boosted without harsh and damaging treatments such as chemo or radiation therapy. Perhaps certain people are at greater risk of cancer development due to abnormal expression of such cancer resistance genes and these could be identified and receive “booster” treatments to increase the fitness of normal cells and thereby increase the detection and elimination of Cancer cells.
The figure to the right indicates how Drosophila genetics can help us address these questions. Genetic tools enable us to knockdown genes in the cells surrounding the RET expressing cells. We can then measure the effect of this to the spread of RET expressing cells.
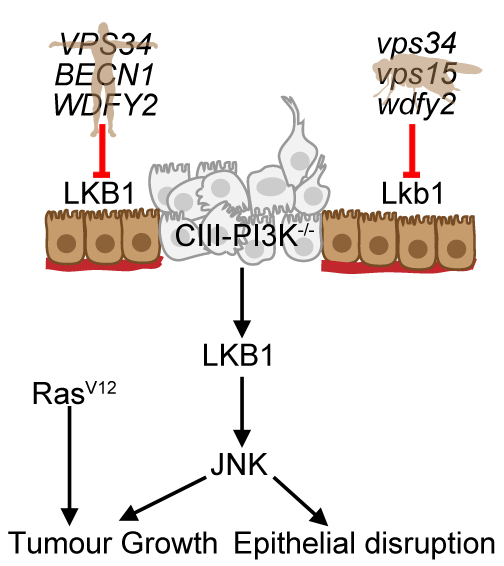
Project 3. Drosophila as a model to study the contextual oncogene LKB1.
LKB1 (Liver Kinase B1) is a contextual oncogene, that is, in certain tumours it promotes growth and survival. It is also thought to be a tumor suppressor and genetic evidence shows it to be responsible for the rare and complex illness, peutz-jeghers syndrome (https://ghr.nlm.nih.gov/condition/peutz-jeghers-syndrome#genes), which inevitably causes death from cancer, typically gastrointestinal. Despite the strong correlation between LKB1 mutations and cancer, how LKB1 mutations actually cause these symptoms in not well understood. We (and others) have shown that the activity of LKB1 is controlled in part by its localisation within the cell via its association with intracellular membranes. I aim to precisely pinpoint the interplay between vesicle trafficking and LKB1 activity. Understanding this could give new inroads into understanding LKB1 cancers and potentially generate new therapeutics for sufferers of Peutz-Jeghers syndrome. With this in mind I have registered myself as a participant within the Rare Diseases Models & Mechanisms – Europe (RDMM-Europe) (http://solve-rd.eu/rdmm-europe/), offering Drosophila as a model for study of this disease. As the human LKB1 gene can substitute for the Drosophila gene, rescuing lethality, Drosophila offers an attractive model to introduce human LKB1 variants, expressing them broadly or narrowly as a restricted field of cells in a wildtype or heterozygote background and investigate the consequences to cells/tissue development.
This work follows on from findings made during recent work, a summary of which is shown below.